GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low
4.8 (371) · $ 21.99 · In stock
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
jpoell · GitHub

GitHub - Nealelab/whole_genome_analysis_pipeline
libtool: error: cannot determine absolute directory name of

PDF) OTSUCNV: an adaptive segmentation and OTSU-based anomaly
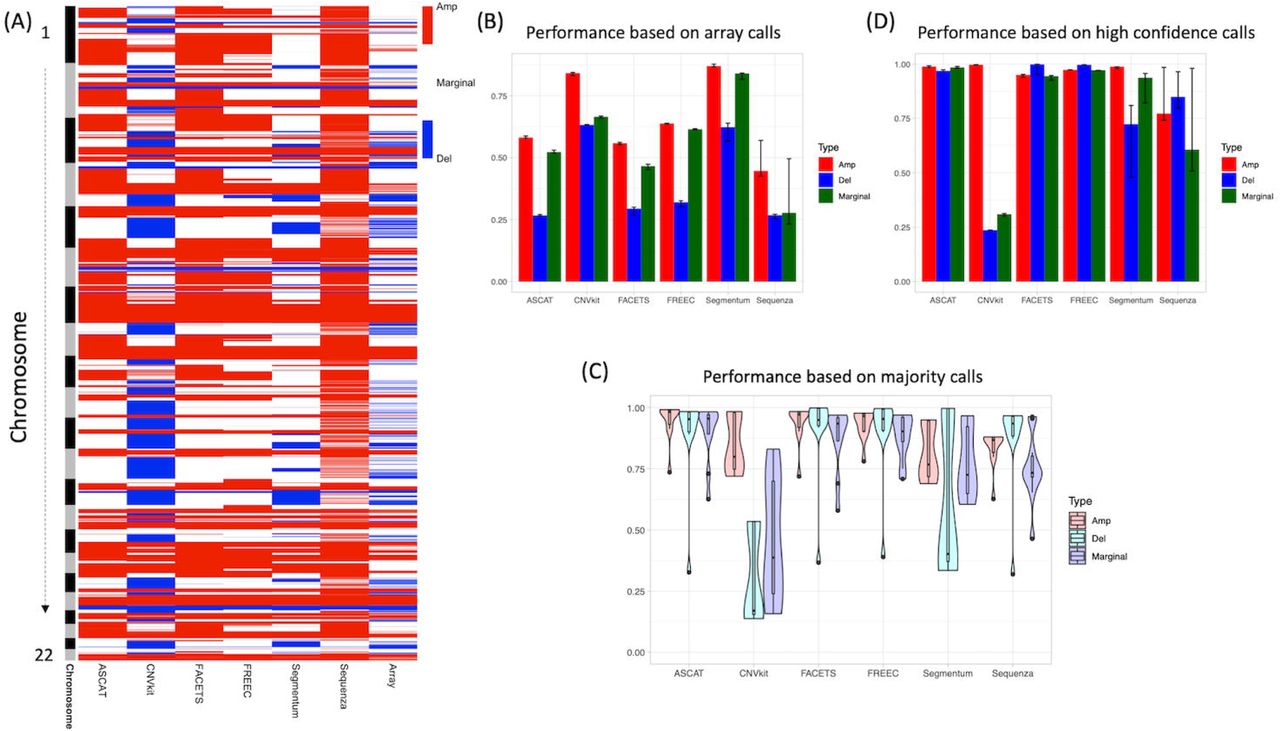
Comprehensive Assessment of Somatic Copy Number Variation Calling

PDF) OTSUCNV: an adaptive segmentation and OTSU-based anomaly

Comprehensive Assessment of Somatic Copy Number Variation Calling
GitHub - wyguo/genotype_specific_RTD: Core scripts of data

Comprehensive Assessment of Somatic Copy Number Variation Calling

PDF) WAVECNV: A New Approach for Detecting Copy Number Variation

American Society for Clinical Pharmacology and Therapeutics - 2019

Absolute quantification reveals the stable transmission of a high

CNApp: copy number alterations integrative analysis

GitHub - Nealelab/whole_genome_analysis_pipeline







