GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
4.9 (360) · $ 9.50 · In stock
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
PDF) Oral cancer prediction by noninvasive genetic screening

Computational validation of clonal and subclonal copy number alterations from bulk tumor sequencing using CNAqc, Genome Biology

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data

CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing, Genome Biology
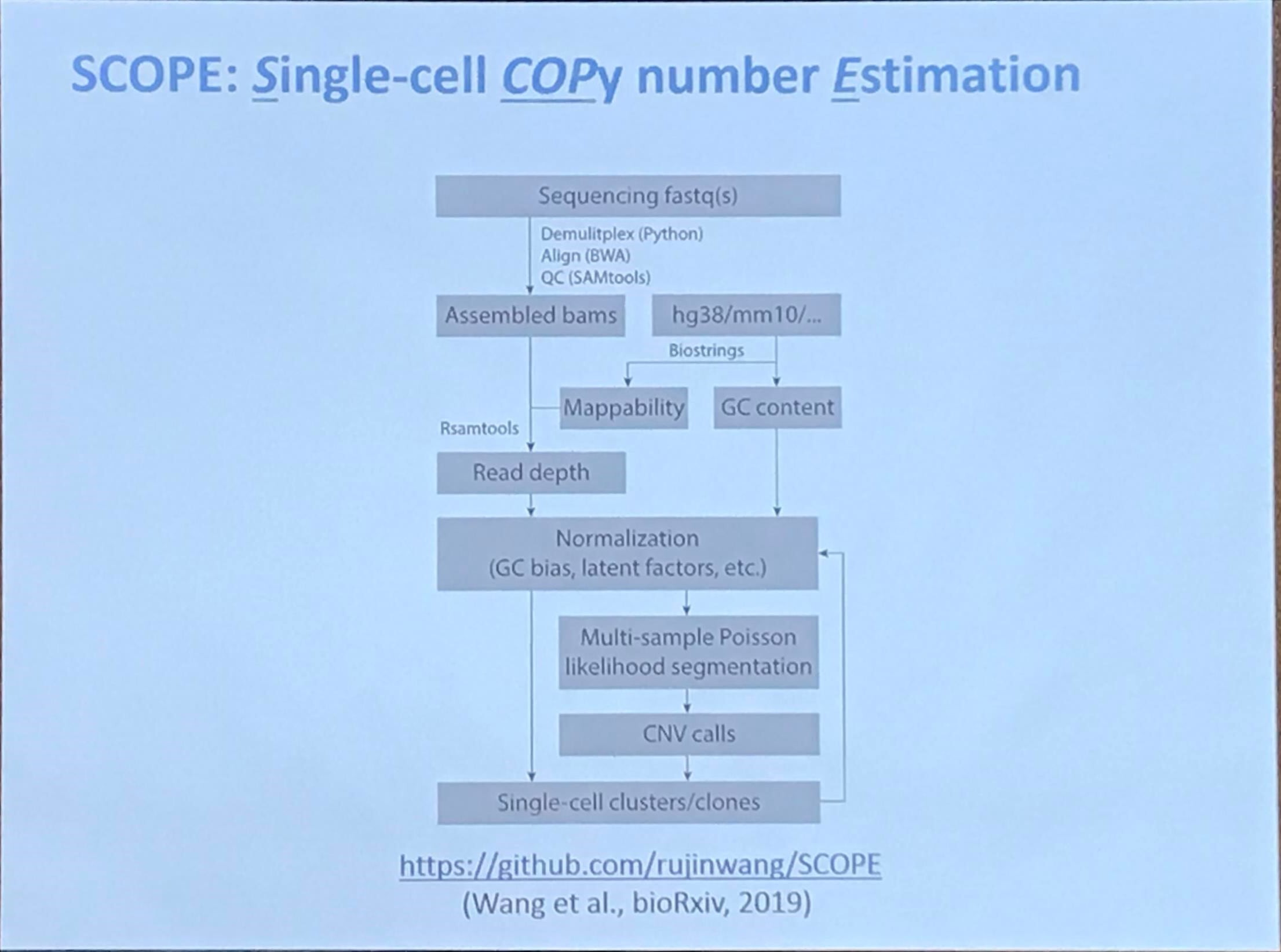
DNA copy number profiling: from bulk tissue to single cells
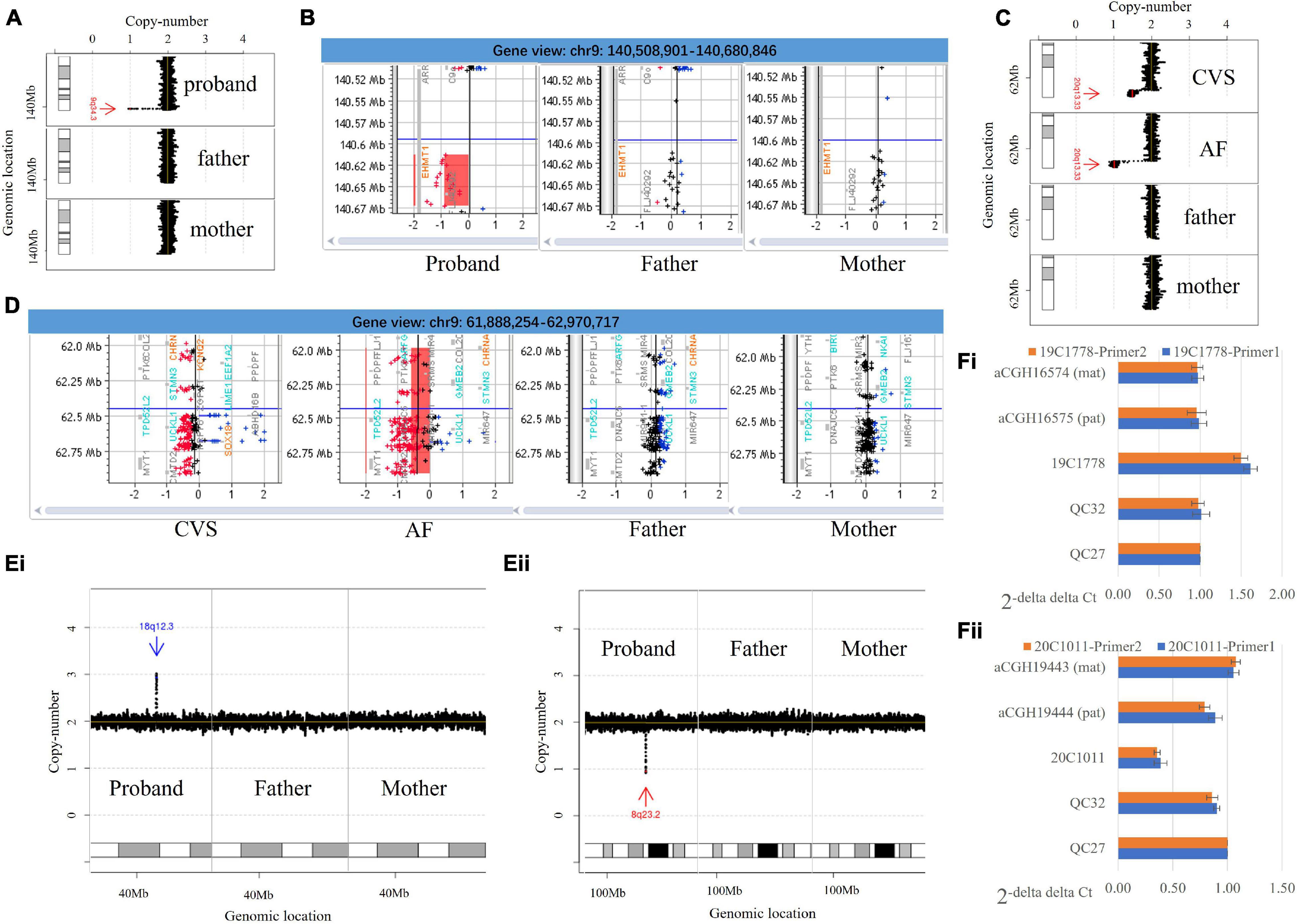
Frontiers Trio-Based Low-Pass Genome Sequencing Reveals Characteristics and Significance of Rare Copy Number Variants in Prenatal Diagnosis

RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data

Ensemble of nucleic acid absolute quantitation modules for copy number variation detection and RNA profiling
GitHub - AdelmanLab/GetGeneAnnotation_GGA
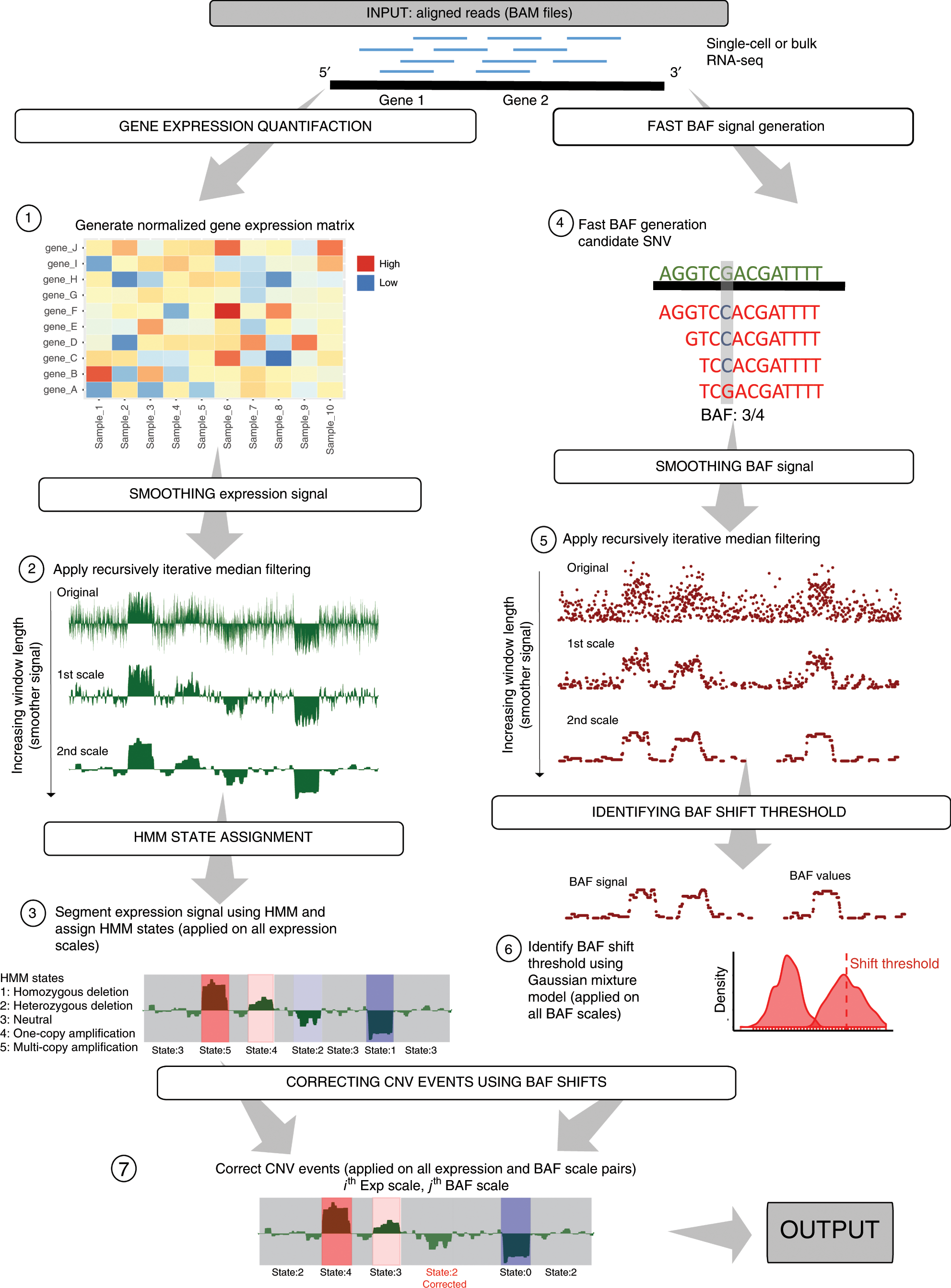
CaSpER identifies and visualizes CNV events by integrative analysis of single-cell or bulk RNA-sequencing data
PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data
jpoell · GitHub

GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data
PDF) OTSUCNV: an adaptive segmentation and OTSU-based anomaly classification method for CNV detection using NGS data

GitHub - Nealelab/whole_genome_analysis_pipeline