SPANDx workflow for analysis of haploid next-generation re
4.8 (165) · $ 18.99 · In stock


Criteria for calling a SNP in each of the four algorithms

Whole genome sequencing for antimicrobial resistance mechanisms, virulence factors and clonality in invasive Streptococcus agalactiae blood culture isolates recovered in Australia - Pathology

Comparative genomics and antimicrobial resistance profiling of Elizabethkingia isolates reveals nosocomial transmission and in vitro susceptibility to fluoroquinolones, tetracyclines and trimethoprim-sulfamethoxazole

Single-Cell RNA Sequencing of Human, Macaque, and Mouse Testes Uncovers Conserved and Divergent Features of Mammalian Spermatogenesis - ScienceDirect
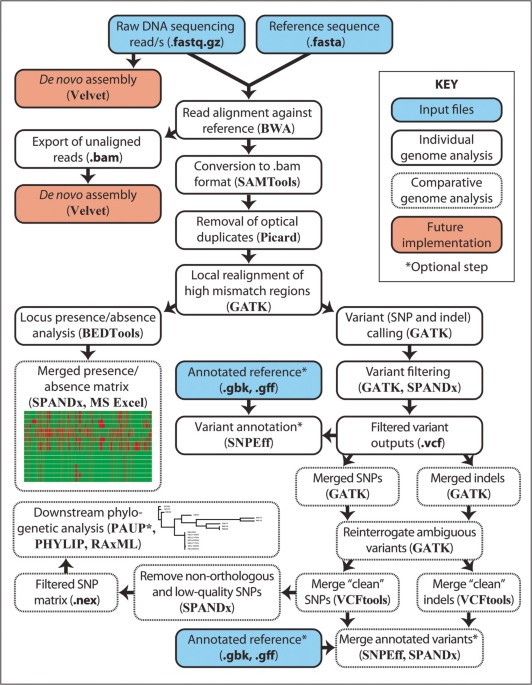
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes
Save time with our One-Day-Workflow - GenDx
![minSNPs: an R package for the derivation of resolution-optimised SNP sets from microbial genomic data [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2023/15339/1/fig-1-2x.jpg)
minSNPs: an R package for the derivation of resolution-optimised SNP sets from microbial genomic data [PeerJ]

SPANDx workflow for analysis of haploid next-generation re-sequencing data.
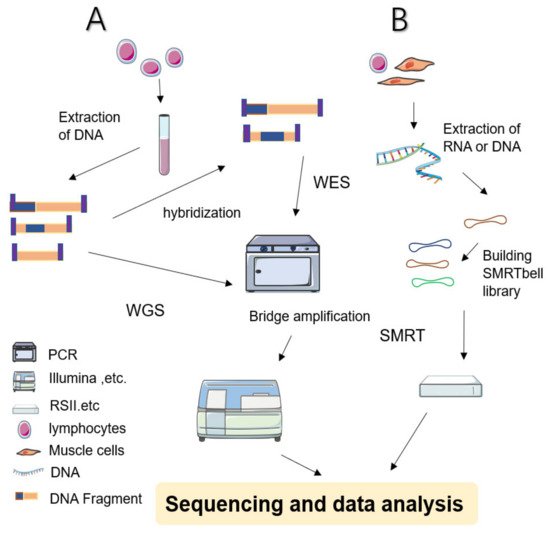
Next-Generation Sequencing in Neurogenetic Diseases

SPANDx workflow for analysis of haploid next-generation re-sequencing data.
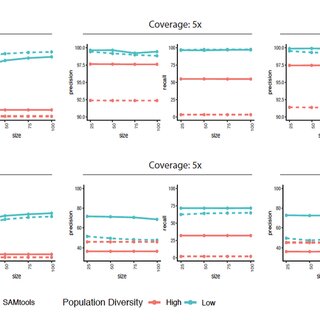
Criteria for calling a SNP in each of the four algorithms
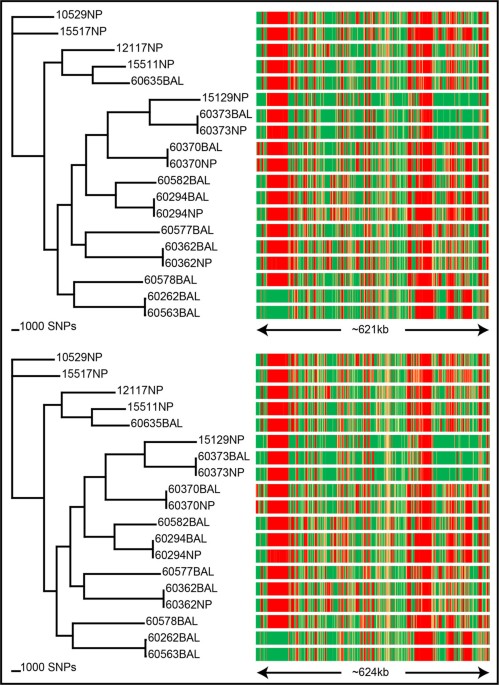
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes

Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of 'Haemophilus intermedius







